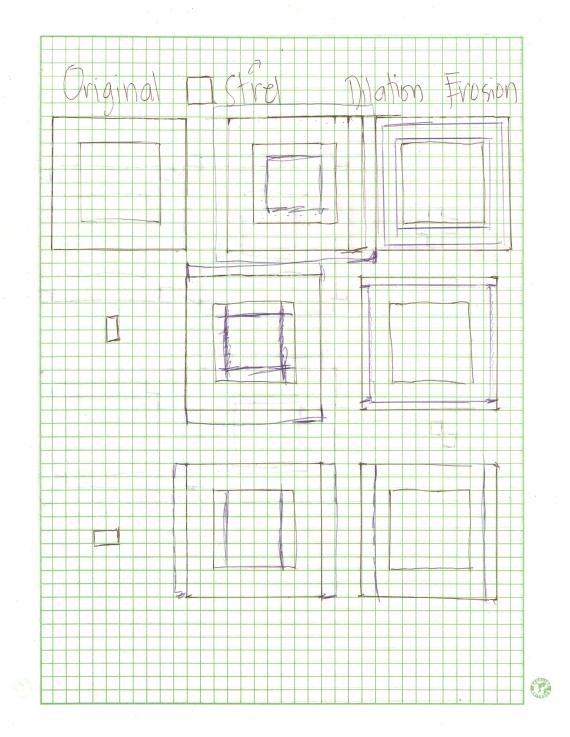
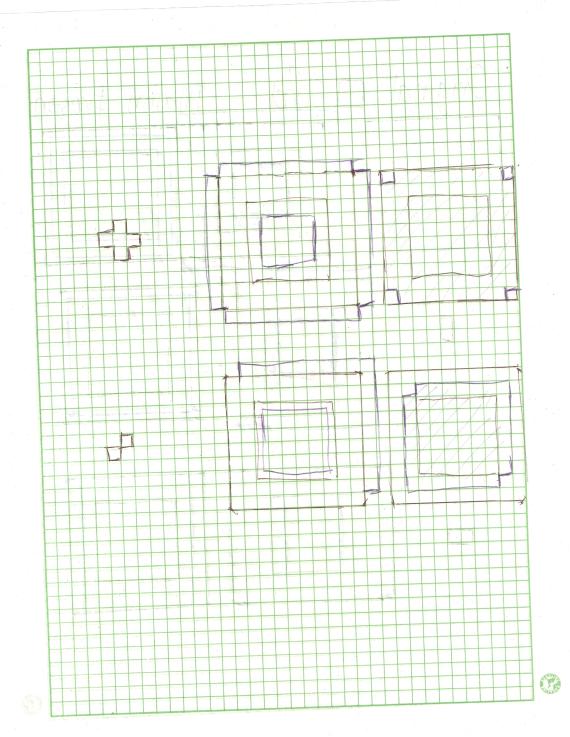
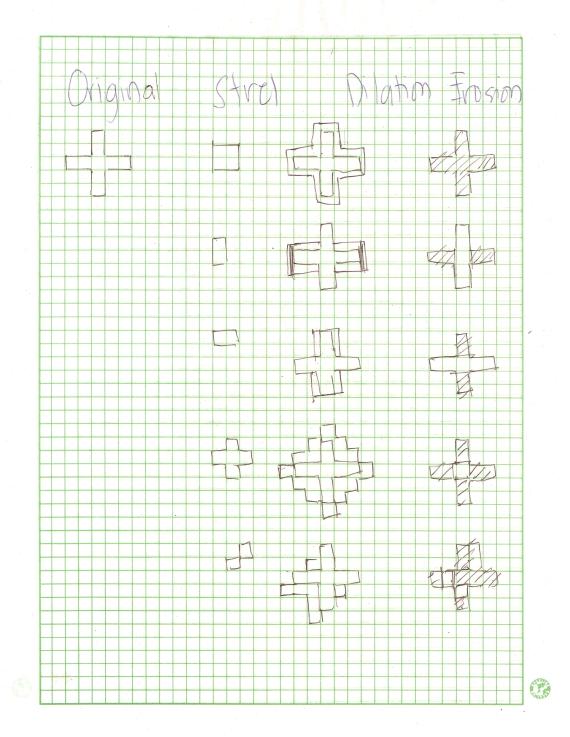
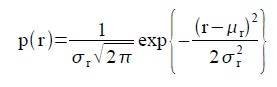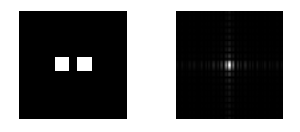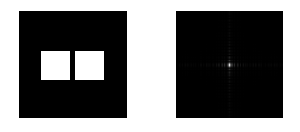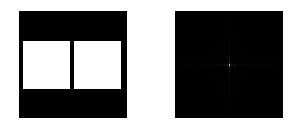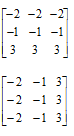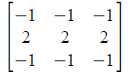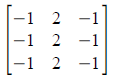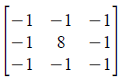Images of low contrast both suffer from their lack of aesthetic value and from supposed concealment of some necessary informations. They can be attributed to many factors such as poor focusing, presence of smoke, low lighting levels during imaging, lens flare [1], etc. Although sometimes they are unavoidable, image enhancement can be done to enhance the contrast and furtheremore retrieve the supposed data loss. Enhancement is a necessity particularly to images used in the practical sciences like in medicine and in astronomy. Conventional image processing programs such as Adobe Photoshop© and its freeware substitute, GIMP offer a lot of options to enhance a low contrast image. One less explored way of image enhancement is through histogram manipulation and our heroic freeware Scilab can once again make a way to perform this operation.
The graylevel Probability Distribution Function (PDF) is equal to the normalized histogram of a grayscale image. In this activity, image enhancement will be performed by modifying a grayscale PDF through backprojection of a desired Cumulative Distribution Function (CDF) to it [2]. We chose to manipulate the PDF through the CDF because it is easier to produce a smooth plot that will have just a slight difference with the original CDF compared to directly manipulating the spectrum-like plot of a PDF. Ideally, CDF’s are not smooth and undefinable using simple functions. Here, four ideal CDFs will be backprojected; a linear and 3 non linear functions to simulate the response of the human eye which is known to be nonlinear. These will include a quarter circle, a parabolic and a sigmoid function defined using a hyperbolic tangent function. The PDF of the enhanced images will be compared with those of the original image. The CDF will be also taken to show that the new images really follow the behavior of the backprojected CDFs. In performing the backprojection, the interp1 command in Scilab was used. Shown below is an image showing the step in altering the grayscale distribution [2].
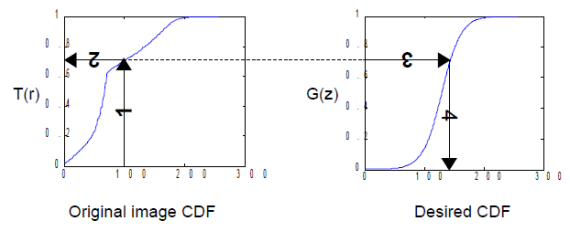
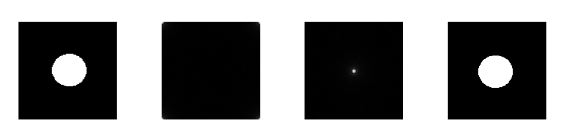
Figure 1. Steps in altering the grayscale distribution.( 1) From pixel grayscale, find CDF value (2). Trace this value in the desired CDF (3). Replace pixel value by grayscale value having this CDF value in desired CDF (4).
For the linear CDF, the equation used is the equation of a line with slope 1/255. For all plots displayed the x range is 0 to 255, and y range is 0 to 1. For the quarter circle, the equation used is just the equation of a circle centered at (255, 0). A simple parabola of order 2 was plotted for the quadratic and a hyperbolic tangent function {tanh(16.*((a-127.5)./255))} was used to plot a desired CDF similar to the one shown in the figure below.
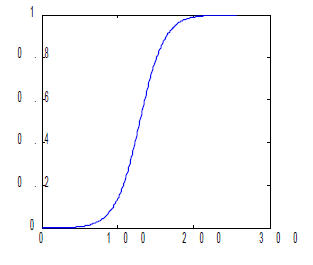
Figure 2. Desired CDF
Shown below are examples of the image enhancement done using these steps, the original image will be displayed together with the enhanced images using the desired CDFs, the 4 CDFs to be backprojected will be also shown, and finally the PDFs and CDFs of the original and the enhanced images will be displayed. To evaluate the capability of the program in image enhancement I will be applying it on 4 images. From these side by side plots, the manifestation of the manipulation can be easily visualized. (I used subplot for multiple plotting, although it made my code painfully long it looks more conclusive that way ).
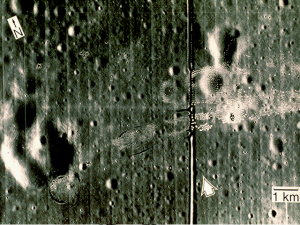
The first image is a personal pic from our hiking last summer in Batangas.

Figure 3. Image to be enhanced
The plot of the ideal CDFs to be backprojected to the original PDF is shown below
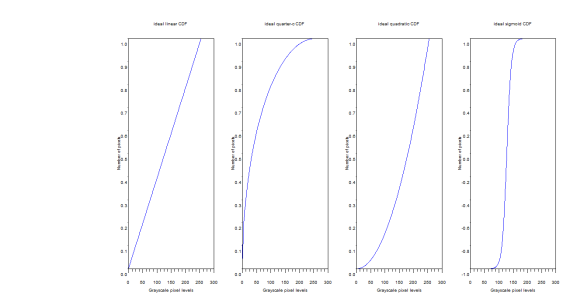
Figure 4. Ideal CDFs to be bacjprojected to the PDF of the original image

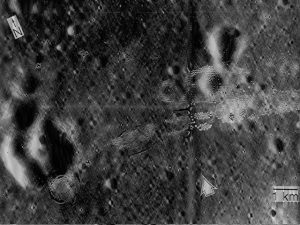
(a) (b) (c) (d) (e) Figure 5. Image Enhancement; (a) Original Image, (b) Enhanced image with Linear CDF, (c) with quarter circle CDF, (d) parabolic CDF, and (e) sigmoid CDF
From these figures we can easily see the effect of backprojecting different CDFs to a grayscale image. However, the choice on the best CDF to use still depends on the application of the image enhancement.
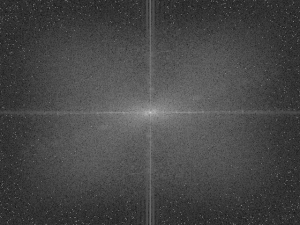
The corresponding PDFs are shown.
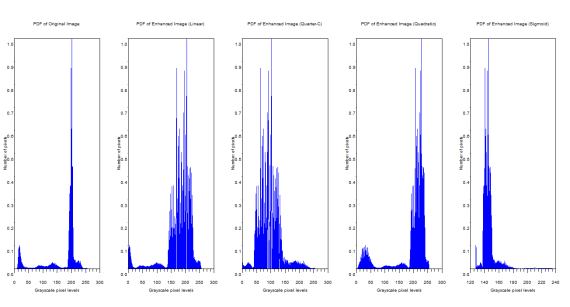
Figure 6. PDFs of the original and enhanced images
The manifestation of the ideal CDF backprojection can be easily seen from the PDFs. The graylevel distributions changed. The original image has grayscale maximum of 255 and a minimum of 0.228. The linear CDF-enhanced image now has a grayscale max of 255 and a min of 0.000659. For the quarter circle the max is 255 and the min is 0.0000291. The quadratic has a max of 255 and a min of 0.1672454. Finally the sigmoid function has a max of 253.603 and a min of 127.59751.
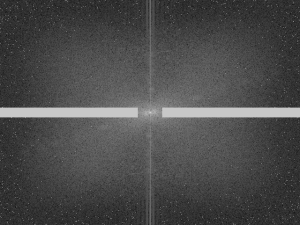
And the corresponding CDFs…
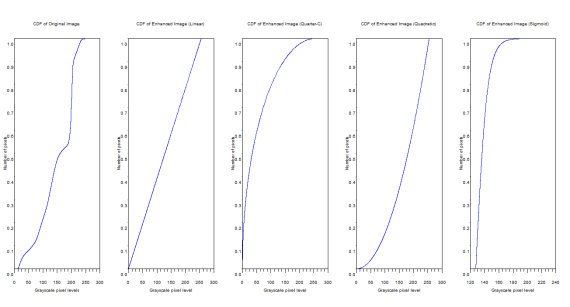
Figure 7. CDFs of the original and enhanced images
From the above figure, it can be inferred that the CDF of the enhanced images really do follow the backprojected ideal CDFs.
I also tried enhancing several images using these codes. Shown below in succession are the figures of the original and the enhanced image, the PDFs and the CDFs.
The image is of a flower

Figure 8. Original and Enhanced Images
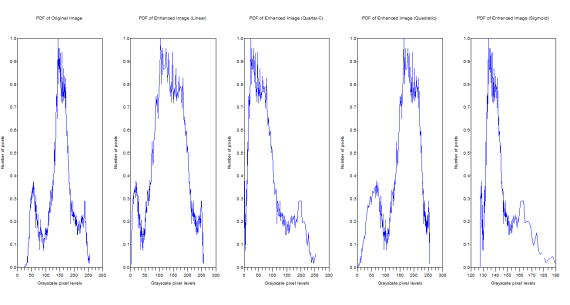
Figure 9. PDFs of the original and the enhanced image
The original image has grayscale maximum of 255 and a minimum of 24. The linear CDF-enhanced image now has a grayscale max of 255 and a min of 0.0153245. For the quarter circle the max is 255 and the min is 0.0006729. The quadratic has a max of 255 and a min of 1.9692508. Finally the sigmoid function has a max of 189.45846 and a min of 127.50096.
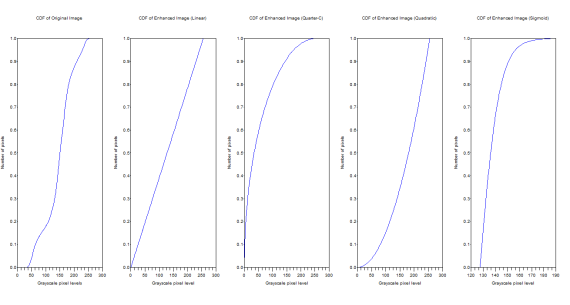
Figure 10. CDFs of the original and the enhanced images
I also tried enhancing another photo taken from our Batangas hiking, this time it is an image of the shore.

Figure 11. Original and Enhanced Images

Figure 12. PDFs of the original and the enhanced images
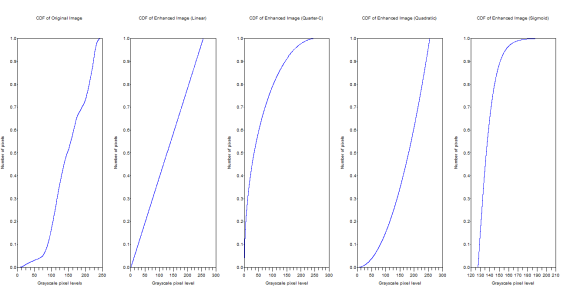
Figure 13. CDFs of the original and the enhanced images
Lastly, I tried the histogram manipulation on the image of the the standard image processing subject, the famous Lena Sjobloom.
Below is her image taken from http://ndevilla.free.fr/lena/

Figure 14. Image of Lena Sjobloom, popular image processing test subject
The grayscale image and the enhanced images are shown below

Figure 15. Original and Enhanced Images
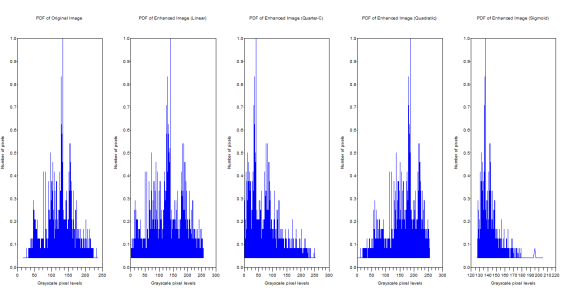
Figure 16. PDFs of the Original and the Enhanced Images
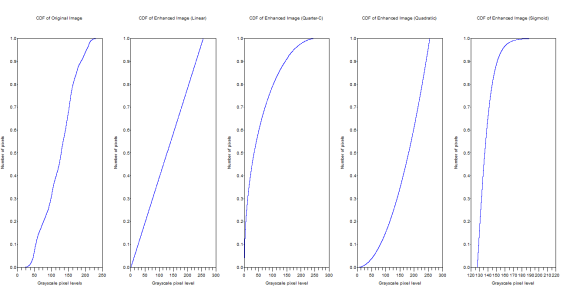
Figure 17. CDFs of the original and enhanced images
I think I have already presented enough examples of the enhancements done using histogram manipulation done on Scilab.
Now let us revert back to conventional image processing. The freeware GIMP has a command curve where the histogram can be manipulated. Shown below is an image of how it is done in GIMP.

Figure 18. Histogram Manipulation in GIMP
Shown below are examples of the enhancements done using different curves.

Figure 19. Enhanced image using Linear Color Curve

Figure 20. Enhanced image using linear (negative slope) color curve

Figure 21. Enhanced Image using quadratic color curve
I give myself a grade of 10 for this activity, being able to do all the necessary things.
Appendix
CODES USED:
chdir(‘C:\Users\Rob H. Entac\Documents\Acads\1st Semester 10-11\App Phy 186\Act 5’);
stacksize(100000000);
I = imread(‘london.jpg’);
GIm = im2gray(I);
scf(0);
subplot(151);
imshow(GIm);
//PDF
GI = GIm*255;
pdf = tabul(GI,’i’);
GI_max = max(GI)
GI_min = min(GI)
//CDF
cdf = cumsum(pdf(:,2));
cdf = cdf/max(cdf);
//CDF ideal linear
a = [0:1:255];
b = [0:1/255:1];
//Backprojection Linear CDF
i = interp1(pdf(:,1),cdf,GI);
j = interp1(b,a,i);
j_max = max(j)
j_min = min(j)
subplot(152);
imshow(j,[0,255]);
//PDF of Enhanced Image (Linear)
Elpdf = tabul(j,’i’);
//CDF of Enhanced Image (Linear)
Elcdf = cumsum(Elpdf(:,2));
Elcdf = Elcdf/max(Elcdf);
//CDF ideal quarter-c
c = sqrt((255)^2 – (a-255)^2);
c = c/max(c);
//Backprojection quarter-c CDF
k = interp1(pdf(:,1),cdf,GI);
l = interp1(c,a,k);
l_max = max(l)
l_min = min(l)
subplot(153);
imshow(l,[0,255]);
//PDF of Enhanced Image (Quarter-C)
Eepdf = tabul(l,’i’);
//CDF of Enhanced Image (Quarter-C)
Eecdf = cumsum(Eepdf(:,2));
Eecdf = Eecdf/max(Eecdf);
//CDF ideal quadratic
d = a^2;
d = d/max(d);
//Backprojection Quadratic
m = interp1(pdf(:,1),cdf,GI);
n = interp1(d,a,m);
n_max = max(n)
n_min = min(n)
subplot(154);
imshow(n,[0,255]);
//PDF of Enhanced Image (Quadratic)
Eqpdf = tabul(n,’i’);
//CDF of Enhanced Image (Quadratic)
Eqcdf = cumsum(Eqpdf(:,2));
Eqcdf = Eqcdf/max(Eqcdf);
//CDF ideal sigmoid
e = tanh(16.*((a-127.5)./255));
//Backprojection Sigmoid
o = interp1(pdf(:,1),cdf,GI);
p = interp1(e,a,o);
p_max = max(p)
p_min = min(p)
subplot(155);
imshow(p,[0,255]);
//PDF of Enhanced Image (Sigmoid)
Espdf = tabul(p,’i’);
//CDF of Enhanced Image (Sigmoid)
Escdf = cumsum(Espdf(:,2));
Escdf = Escdf/max(Escdf);
//PLOTTINGcodes
//PDF original
scf(1);
subplot(151);
plot(pdf(:,1),pdf(:,2)/max(pdf(:,2)));
title(‘PDF of Original Image’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//PDF linear enhanced
subplot(152);
plot(Elpdf(:,1),Elpdf(:,2)/max(Elpdf(:,2)));
title(‘PDF of Enhanced Image (Linear)’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//PDF quarter-c enhanced
subplot(153);
plot(Eepdf(:,1),Eepdf(:,2)/max(Eepdf(:,2)));
title(‘PDF of Enhanced Image (Quarter-C)’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//PDF quadratic enhanced
subplot(154);
plot(Eqpdf(:,1),Eqpdf(:,2)/max(Eqpdf(:,2)));
title(‘PDF of Enhanced Image (Quadratic)’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//PDF sigmoid enhanced
subplot(155);
plot(Espdf(:,1),Espdf(:,2)/max(Espdf(:,2)));
title(‘PDF of Enhanced Image (Sigmoid)’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//CDF original
scf(2);
subplot(151);
plot(pdf(:,1),cdf);
title(‘CDF of Original Image’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//CDF linear
subplot(152);
plot(Elpdf(:,1),Elcdf);
title(‘CDF of Enhanced Image (Linear)’);
xlabel(‘Grayscale pixel level’);
ylabel(‘Number of pixels’);
//CDF quarter-c
subplot(153);
plot(Eepdf(:,1),Eecdf);
title(‘CDF of Enhanced Image (Quarter-C)’);
xlabel(‘Grayscale pixel level’);
ylabel(‘Number of pixels’);
//CDF quadratic
subplot(154);
plot(Eqpdf(:,1),Eqcdf);
title(‘CDF of Enhanced Image (Quadratic)’);
xlabel(‘Grayscale pixel level’);
ylabel(‘Number of pixels’);
//CDF sigmoid
subplot(155);
plot(Espdf(:,1),Escdf);
title(‘CDF of Enhanced Image (Sigmoid)’);
xlabel(‘Grayscale pixel level’);
ylabel(‘Number of pixels’);
//ideal linear CDF
scf(3);
subplot(152);
plot(a,b);
title(‘ideal linear CDF’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//ideal quarter-c CDF
subplot(153);
plot(a,c);
title(‘ideal quarter-c CDF’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//ideal quadratic CDF
subplot(154);
plot(a,d);
title(‘ideal quadratic CDF’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
//ideal sigmoid CDF
subplot(155);
plot(a,e);
title(‘ideal sigmoid CDF’);
xlabel(‘Grayscale pixel levels’);
ylabel(‘Number of pixels’);
References:
1. Low Contrast Images. <http://en.mimi.hu/photography/low_contrast.html>
2. M. Soriano. Enhancement by Histogram Manipulation. Applied Physics 186 Activity 5 Manual. 2010.